Objectives
Upon completion of this module topic, you should:
- be able to set up a Polymerase Chain Reaction (PCR) using DNA samples, primers, buffer, dNTPs, and polymerase enzyme;
- understand the significance and role of each component in the reaction;
- be able to load PCR samples into a thermocycler machine under supervision and understand the mechanisms behind the different steps taking place inside of the thermocycler throughout the PCR reaction;
- have a general understanding of Real-Time PCR and its applications for quantitating gene expression;
- understand the mechanism of DNA sequencing and the methods implemented for sequencing DNA;
Part a
Role of Restriction Enzymes & Enzyme Digestion
This is Part A, Role of Restriction Enzymes & Enzyme Digestion, under the module topic, DNA Restriction & Nucleic Acid Analysis. This topic part has two sections Content Tutorial, and Animations.
Content Tutorial
University of Calgary Biotechnology Training Centre
Restriction Enzymes and Cloning
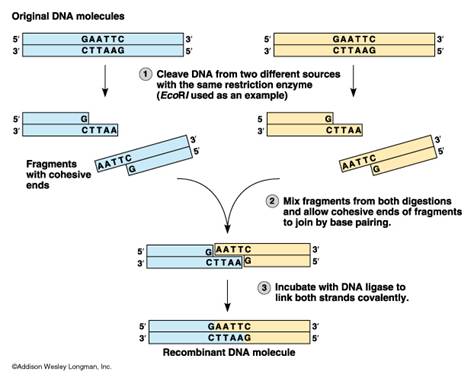
This section on cloning is included with the basics module as it is a necessary basic for research laboratories, but may be required from time to time in clinical labs when something really interesting is found in the molecular lab. Cloning of a gene sequence (or part) is also required to have stable templates for the production of probes, or to save that interesting PCR product. Long term storage of PCR products without cloning them, leaves them susceptible to degradation by bacterial exonucleases (enzymes that digest DNA from the ends). Restriction enzymes are the backbone reagents of cloning, but are used in clinical applications associated with fingerprinting – genetic identity, epidemiology, and in preparation for blotting for other applications. Cloning Step 1: Restriction digestion. This is a step that essentially cuts DNA into little bits. We will see later the application of this to identification (DNA fingerprinting). The molecular scissors are called restriction endonucleases. Restriction endonucleases are enzymes that cleave DNA. They are derived from bacteria where they function in cleaving foreign DNA and thus protecting the integrity of the host bacteria (a bacterial immune system). These enzymes recognize specific base sequences in double stranded DNA. The base sequence serves as a code for the restriction enzyme saying “cut here”. For example, the enzyme labelled EcoRI recognizes the GAATTC sequence of bases and cuts between the G and A.


The sites on the DNA molecule recognized by the enzymes are called restriction sites. Restrictions sites are palindromic, reading the same 5’?3’ on either strand of the DNA. The various enzymes are named for the bacteria from which they have been derived. Eco is derived from Escherichia coli and Hin from Haemophilus influenzae, for example. You will note that the first panel above the cut leaves no overhang and is called a blunt cut leaving blunt ends. Any blunt end can be rejoined to any other blunt end – something to remember when cloning PCR products. The other panel shows a cut that leaves overhanging ends called either 5’ or 3’ overhangs depending on which strand is overhanging. These overhanging ends are also called “sticky ends”.
Enzymes may be chosen that will cleave on either side of a desired DNA sequence. In these cases, cut sites have been predetermined by finding the sequence of nucleotides that encode a specific protein or characteristic. In other cases, various endonucleases will be tried.
Once the DNA has undergone restriction digestion, it may be used to recombine with any other piece of DNA that has the complementary ends, regardless of the source of that DNA. This allows insertion of a segment of DNA into a plasmid that has been cleaved with the same enzyme(s).
Animations
1. Follow the link below to the DNA Interactive Website. – Click on “Manipulation” and then click on the “Techniques” module followed by clicking on the “Cutting & Pasting” section.
DNA Interactive Website (www.dnai.com, Multimedia Page)
a. Task: View the two animations provided: Cutting and Pasting DNA (2D) and Recombining DNA (3D) to learn about how restriction enzymes function and how they are used as an essential tool in recombinant DNA technology.
b. Task: View the video Interviews provided on the website, their titles are listed below:
- First recombinant DNA
- Utility of recombinant DNA
- On the phenomenon of restriction
- Rearranging plasmid DNA
2. Restriction Endonucleases – Animation from Raven, Johnson, Losos, & Singer, Biology, 7th Edition, 2005 Click on the following link to view the animation list.
Restriction Endonucleases (highered.mcgraw-hill.com, Multimedia Page)
Click on the animation titled “Restriction Endonucleases” to learn about restriction enzymes, their functions and their applications to molecular cloning and making recombinant DNA.
Part b
Nucleic Acid Analysis
This is Part B Nucleic Acid Analysis, under the module topic, DNA Restriction & Nucleic Acid Analysis. This topic part has three sections: Content Tutorial, Animations and Activities.
Content Tutorial
Pre-Detection: Electrophoresis Techniques
Usually prior to blotting the DNA, RNA, or protein molecules for further studies and detection, nucleic acids are separated based on size or mass by electrophoresis. Like total protein electrophoresis, DNA and RNA can be separated by size/mass, but charge is irrelevant (DNA and RNA are negatively charged). In protein electrophoresis for molecular biology, proteins are separated by mass as charge is made negative by the pH of the buffer used. (This technique will be explored in module III). As there are no issues with charge, the only difference with the molecules position in the matrix following electrophoresis is the length of the sequence. Thus it is possible to determine the length of a piece of DNA or RNA or mass of a protein by its position in a gel relative to a standard marker. This type of analysis is important in epidemiology, forensics and in just checking up on your PCR reaction. For proteins many techniques are available to determine function as well as mass. Two major types of matrix are used for electrophoresis; agarose gel or acrylamide gel. Agarose is of superior optical clarity compared to microbiology agar, but is handled in similar ways. Acrylamide is a crosslinked polymer that enables the use of small amounts of sample, separation of very similar pieces (sequencing gels for example) and are able to take a great deal of heat during the electrophoresis run. Buffers are added to the gels when they are made. DNA and RNA can be visualized in the gels after electrophoresis by the addition of a fluorescent dye that can be seen under UV light. Usually pictures or digital images are used for determination of the size of the bands in the gel.
Agarose Gel Electrophoresis
Electrophoresis is a method whereby charged molecules in solution migrate in response to an electric field. Because of the phosphate backbone in DNA molecules, they have a net negative charge and migrate toward the anode (positive pole) in an electric field. Their rate of migration or mobility is related to the strength of the field, the size of the molecule, as well as the medium (gel) in which they are migrating. For separation of DNA molecules, electrophoresis is often carried out in a horizontal apparatus containing a gel made of agarose. Agarose is a highly purified polysaccharide derived from agar (seaweed) that is not contaminated with charged material. It comes in powder form and is dissolved by boiling in aqueous solutions. It remains in a liquid state until the temperature is lowered to about 40ºC at which point the agar solution gels. The gel is stable and will not dissolve again until raised back to boiling temperatures. The pore size of the agarose is adjusted by changing the concentration of the agarose in the gel. The higher the concentration of agarose, the smaller the gel pore size. Working concentrations are usually in the range of 0.5% to 2.0%. A 2.0% gel has a smaller pore size than a 0.5% gel and will allow better separation of short DNA molecules (those that are a few hundred base pairs in length) than a 1.0% gel which might be used to separate molecules that are larger in size (thousands of base pairs in length). The migration of molecules in agarose is size-dependent and allows separation of molecules up to about 20 kilobases (kb) in size. The smaller the molecule, the more rapidly it will be able to pass through the pores in the agarose in its migration toward the positive pole. Thus, in a mixture of DNA molecules of different sizes, the shortest fragments will migrate the most rapidly, while the largest will be retarded to the greatest extent by the pores in the agarose and will migrate the most slowly. The best separation is achieved after experimenting with variations in the concentration of agarose and the separation time in the electric field. When the agarose gel is prepared, a “slot maker” or comb is inserted into the chamber so that small wells or slots are formed when the agarose gels or sets. The DNA sample to be analyzed is first mixed with a glycerol-dye solution. The glycerol in the sample makes it more dense than the running buffer so that it can be applied into the sample slot of a submerged gel in an electrophoretic chamber without diffusing away prior to application of electrical current. The inclusion of the dye (bromophenol blue or others) helps to make the sample visible during application and provides a marker to help track the progress of the electrophoretic run because it is also negatively charged and migrates toward the positive pole at a rate similar to that of small DNA molecules. To better visualize the separation of DNA molecules during and after the electrophoretic run, ethidium bromide, SYBR green or SYBR gold are included in low concentrations (0.5 μg/ml) in the gel and in the running buffer, or in the loading dye itself (SYBRs). SYBRs bind to double-stranded DNA and becomes fluorescent when excited by ultraviolet (UV) light. A DNA ladder that contains DNA molecules of defined length is also included as one of the samples in one of the lanes of the gel to provide an internal marker of DNA fragment sizes and to provide an indication of how well molecules have separated in the gel. The DNA bands can be visualized and photographed by placing the agarose gel on a UV light box. Care must be taken to wear a UV resistant shield, goggles or glasses when viewing gels on a UV box to avoid damage to your eyes. In addition, ethidium bromide is a carcinogen and mutagen and care must be taken to avoid skin contact with ethidium bromide- containing solutions. Always wear latex gloves when working with DNA samples during electrophoretic procedures to prevent contact with ethidium bromide in the gel or running buffer. Other dyes include SYBR green or gold, which are much safer to use and have similar binding characteristics as ethidium bromide, but in some circumstances, are not quite as sensitive in gels. To confirm the identity of a PCR or RT-PCR product, the agarose gel from above can be subjected to the same non-amplification techniques (blotting) as will be presented in Module 4.
Acrylamide Gel Electrophoresis
In some instances, higher resolution gels are required as for separation of small DNA fragments, ssDNA fragments or in manual sequencing, and this can be done using the cross-linked polymer acrylamide. Extreme sensitivity is obtained by staining acrylamide gels with a silver stain. Such gels can become permanent records as well by drying the gel between sheets of dialysis membrane or onto filter paper. The addition of urea to acrylamide gels will ensure that ssDNA (single-stranded DNA) runs according to size rather than shape due to base-pairing within a strand (secondary structure like tRNA). We will explore the applications of acrylamide gel electrophoresis in greater detail in Module III. Protein Techniques. The agarose gel electrophoresis technique can also be used to analyze Polymerase Chain Reaction (PCR) products which will be discussed in detail in module subset II-b. Module subset II-c. and II-e., will present information on further applications for using an agarose gel for nucleic acid hybridization (blotting) techniques as well as DNA (gel) extraction techniques.
- Agarose Gel Electrophoresis Procedure Pictures (PDF Document)
- Powerpoint: View the Agarose Gel Electrophoresis Slides (PowerPoint Document)
RFLP or DNA Fingerprinting
Prior to electrophoresis, an additional step can be performed that essentially cuts the whole genomic DNA into little bits for fingerprinting or cutting PCR products into smaller pieces. The molecular scissors are called restriction endonucleases. Restriction endonucleases are enzymes that cleave DNA. They are derived from bacteria where they function in cleaving foreign DNA and thus protecting the integrity of the host bacteria. These enzymes recognize specific base sequences in double stranded DNA. The base sequence serves as a code for the restriction enzyme saying “cut here”. For example, the enzyme called EcoRI recognizes the GAATTC sequence of bases and cuts between the G and A.


AWLInc The sites on the DNA molecule recognized by the enzymes are called restriction sites. The various enzymes are named for the bacteria from which they have been derived. Eco is derived from Escherichia coli and Hin from Haemophilus influenzae.

Enzymes may be chosen that will cleave on either side of a desired DNA sequence or at the site of a known/common mutation. In these cases, cut-sites have been predetermined by finding the sequence of nucleotides that encode a specific protein or characteristic mutation. In other cases, various endonucleases will be tried to obtain the desired resolution of a fingerprint; generally 5-40 bands. RFLP is commonly used in epidemiology, mutation detection in genetics and in some forms of HLA typing. One thing about RFLP – it may be followed by gel electrophoresis and staining with ethidium bromide exactly the same as a post-PCR gel would be done, and then the gel is recorded by taking a picture. Gel documentation systems that use band-finding software are able to analyze the bands for size and pattern. The banding patterns can then be used for relationship or phylogenetic analysis.
- Powerpoint: View the DNA Fingerprinting Slides (PowerPoint Presentation)
Animations
1. Sorting and Sequencing– Follow the link below to the DNA Interactive Website. Click on the “Manipulation” section and then click on the “Techniques” module. Continue by clicking on the “Sorting & Sequencing” section (you will focus on the “sorting” content for this module and then you will revisit the website to view the “sequencing” content in module subset II.b.) Task: View the first 2D animation provided: Gel Electrophoresis, to learn about the mechanism of gel electrophoresis. You will view the sequencing animations and interview section in the next module.
- Gel Electorphoresis (www.dnai.org, Multimedia Page)
2. Restriction Fragment Length Polymorphisms (RFLPs) Animations from Raven, Johnson, Losos, & Singer, Biology, 7th Edition, 2005 Click on the following link to again view the animation list. Now click on the animation titled “Restriction Fragment Polymorphisms”. This animation depicts how restriction enzymes can cut DNA at specific sites, producing fragments of DNA at varying lengths that can be analyzed by separating the digested DNA by agarose gel electrophoresis and analyzing the varying fragment sizes.
- Gel Electorphoresis (www.dnai.org, Multimedia Page)
Activities
1. Virtual Lab 1: DNA Electrophoresis The Biology Place Click on the follow link to complete a virtual lab activity. In the virtual lab you will apply the gel electrophoresis technique to samples of DNA that you have been provided with. The DNA samples have been cut by restriction enzymes through a restriction enzyme digestion reaction. Using gel electrophoresis you will separate the fragments and analyze your results by comparing your unknown sample sizes to known standard sizes to assist in calculating the sizes of the unknown samples. You will complete the second part of this virtual lab now and you will complete the first part later on in Module Subset II-d.4. Lab Bench Activity: Molecular Biology Lab 6-II DNA Electrophoresis
- DNA Electrophoresis (www.phschool.com, HTML Page)
Task: Complete Lab Bench Activity, Analysis of Results, and L
2. Virtual Lab 2: Agarose Gel Electrophoresis of Restriction Fragments Colorado State University First click on the link below to access tutorials on Restriction Mapping and Gel Electrophoresis background information before completing the lab.
- Restriction Mapping (www.vivo.colostate.edu, Multimedia Page)
- Gel Electorphoresis (www.vivo.colostate.edu, Multimedia Page)
When you have finished the background section, click on the following link to access the virtual lab.
- Virtual Lab (www.vivo.colostate.edu, Multimedia Page)
3. Virtual Lab 3: Gel Electrophoresis The University of Utah Genetic Science Learning Center For additional practice click on the following link to access the lab.
- Gel Electrophoresis Virtual Lab (learn.genetics.utah.edu, Multimedia Page)
4. Case Study: RFLP’s & DNA Fingerprinting The University of Calgary
Case 1: Screening for the sickle-cell gene
Sickle cell anemia is a genetic disease in which both genes in the patient encode the amino acid valine (Val) in the sixth position of the beta chain (betaS) of the hemoglobin molecule. “Normal” beta chains (betaA) have glutamic acid at this position. The only difference between the two genes is the substitution of a T for an A in the middle position of codon 6. This converts a GAG codon (for Glu) to a GTG codon for Val, and abolishes a sequence (CTGAGG, which spans codons 5, 6, and 7) recognized and cut by one of the restriction enzymes.

Here is the PCR-RFLP gel results for a family. Can you label the pedigree whether with who is a carrier, normal or afflicted individual?
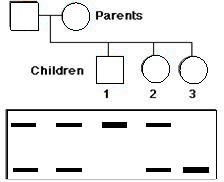
Answer link (users.rcn.com, HTML Page)
Case 2: RFLP DNA Fingerprinting – The image of an autoradiograph (courtesy of Lifecodes Corporation)shows the test results in a rape case. Two probes were used: one revealing the bands at the top, the other those at the bottom.

DNA was tested from
- semen removed from the vagina of the rape victim (EVIDENCE #2)
- a semen stain left on the victim’s clothing (EVIDENCE #1)
- the DNA of the victim herself (VICTIM) to be sure that the DNA didn’t come from her cells
- DNA from two suspects (SUSPECT #1, SUSPECT #2)
- a set of DNA fragments of known and decreasing length (MARKER). They provide a built-in ruler for measuring the exact distance that each fragment travels.
- the cells of a previously-tested person to be sure the probes are performing properly (CONTROL)
One the basis of this test, suspect #2 can clearly be ruled out. None of his bands matches the bands found in the semen. Is suspect #1 guilty? Why or why not? What can you do to prove your point.
Answer link (users.rcn.com, HTML Page)
ALL RIGHTS RESERVED: This material may note be reproduced in whole or part without written permission from the Biotechnology Training Centre, University of Calgary, 3330 Hospital Dr. NW, Calgary, Alberta T2N 4N1
5. Case Study: Genetics of Resistance to HIV Infection Annenberg Rediscovering Biology Click on the following website to complete the case study which explores the association between cells, viruses (specifically HIV), and the human immune system. The case study focuses on the molecular techniques of the Polymerase Chain Reaction and Gel Electrophoresis and demonstrates their real-life medical applications for studying human genetics and diseases.(Annenberg, Rediscovering Biology)
- HIV and AIDS Video (www.learner.org, Multimedia Page)
- HIV and AIDS Online Text (www.learner.org, Multimedia Page)
- Human Evolution Online Text (www.learner.org, Multimedia Page)
- Genomics Video (www.learner.org, Multimedia Page)
- Genomics Online Text (www.learner.org, Multimedia Page)
In addition, you may want to refer to the University of Calgary content tutorial in the next module subset, II-b. to read about and/or review the concept and process of the polymerase chain reaction (PCR). When you have finished reviewing recommended the supplemental materials, click on the following website to view the case study homepage and click on the green “GO” arrow to “Launch Case Study”. (Both “Flash” and “HTML” versions are available on the website for viewing)
- Case Study Link (www.learner.org, Multimedia Page)